

Single nucleus methylomes: See the project site at http://brainome.org
By profiling DNA methylation in human and mouse frontal cortex, we identified differentially methylated regions (DMRs) at which the methylation of CG positions (denoted mCG/CG) differ among cell types (neurons vs. glia) or between young and old (fetal vs. adult). Details of statistical methods for calling DMRs may be found in the supplemental material.
Download BED files containing coordinates of all DMRs. Note that these files refer to the mm9 and hg18 genome references.
CellSort: Automated cell sorting for dynamic fluorescence microscopy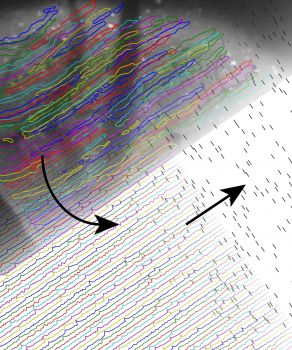 Optical fluorescence microscopy allows neuroscientists to observe and record electrical and chemical activity of brain cells in behaving animals. Techniques such as calcium-dependent fluorscence imaging generate movie data sets, in which information about neural and glial activity is distributed across many pixels and time frames. CellSort is a computational procedure designed to efficiently identify cellular signals by finding combinations of pixels with statistically independent dynamics. The algorithm combines principal and independent component analysis (PCA, ICA) as well as image segmentation and temporal deconvolution.
The image at left illustrates the results of our cell sorting procedure, which estimates cellular spatial filters (top left), calcium-dependent fluorescence dynamics (bottom left), as well as the underlying spike trains (bottom right). The movie (below) shows fluorescence imaging of calcium dynamics in dendrites of mouse cerebellar Purkinje neurons, recorded by in vivo two-photon microscopy. Left panel: Raw data, normalized by the average fluorescence in each pixel (ΔF/F). Right panel: Action potentials within each cell are detected by CellSort. The soundtrack was generated by assigning each dendrite to a different piano key, arranged from the low (upper left) to the high notes (lower right).
|
|
|
|
DeconSTORM: Analysis of Super-Resolution Fluorescence Microscopy by Statistical Deconvolution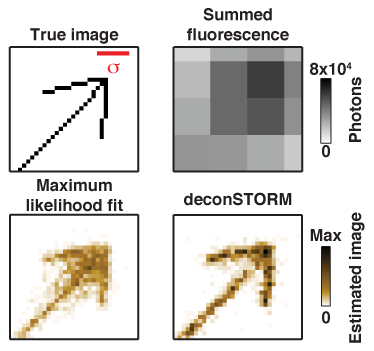 DeconSTORM is a tool for analysis of super-resolution microscopy data sets from of stochastic optical reconstruction micrscopy (STORM), photoactivated localization microscopy (PALM), and related techniques. Our computational analysis approach is described in this publication, and a MATLAB software toolbox is available for implementing our algorithm. |
|
|
Copyright © 2014 - All Rights Reserved - brainome.ucsd.edu
Template by OS Templates